Harnessing Machine Learning to Predict and Analyze Diabetes Risk : A Project Overview
Table of contents
Introduction
Diabetes mellitus is a significant global health challenge, affecting millions of people every year. Early detection and prediction are critical for timely intervention and effective management. This project leverages machine learning (ML) to predict diabetes outcome based on patient data, providing valuable insights into key risk factors and demonstrating the potential of data-driven healthcare solutions. The primary focus of the project is to develop a machine learning model to predict diabetes risk with high accuracy, F1 score and Recall of 80% and secondly, to analyze data to identify significant predictors of diabetes.
To provide a better understanding of diabetes, I have created two concise posts:
Post 1: Discusses diabetes, including its prevalence, symptoms, causes, types, and risk factors.
Post 2: Explores testing methods, prevention strategies, and management approaches.
In this article, I will guide you through the entire process of building this machine learning model, from data loading to preprocessing to model results and evaluation. I will explain the reasoning behind each step and share insights from the results. The notebook is available here
Workflow
This project followed a structured workflow to ensure clarity, efficiency, and robust outcomes. Starting with data exploration and preprocessing, we carefully prepared the dataset by handling missing values, and scaling features. Next, we built and evaluated multiple machine learning models, comparing their performance based on key metrics. Finally, we identified the best-performing model and analyzed feature importance to draw meaningful insights. Find below the workflow outline.
Importing necessary tools
Getting and Loading Data
Data Overview
Data Cleaning
EDA and Feature Engineering
Data Preprocessing
Model Development and Training
Model Evaluation
Conclusion and Reporting
1. Import the necessary tools
# Data analysis tools
import pandas as pd
import numpy as np
# Visualization tools
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
from pandas.plotting import scatter_matrix
# Training machine learning models
from sklearn.preprocessing import RobustScaler
from sklearn.model_selection import train_test_split, cross_val_predict, cross_val_score
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report, precision_score, recall_score, f1_score
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import GradientBoostingClassifier, RandomForestClassifier
from sklearn.svm import SVC
from sklearn.naive_bayes import GaussianNB
from sklearn.tree import DecisionTreeClassifier
#mis
%matplotlib inline
plt.style.use('ggplot')
In this section, we import the essential tools and libraries needed for data analysis, visualization, and machine learning model training.
Data Analysis Tools: Libraries like
pandasandnumpyare used for data manipulation and numerical computations.Visualization Tools:
matplotlibandseabornhelp create informative visualizations to explore patterns and trends in the data. The%matplotlib inlinecommand ensures plots are displayed directly within Jupyter notebooks.Machine Learning Tools: Various modules from
sklearnenable preprocessing, training, and evaluation of models.Default Plot Style: Setting
plt.style.use('ggplot')gives all plots a clean, professional aesthetic.
These imports set up a comprehensive environment to streamline the workflow for building and analyzing machine learning models.
2. Getting and Loading Data
Data is the foundation of any machine learning model, as it is the cornerstone of the success and performance of any machine learning project. The data used for this project is originally from the National Institute of Diabetes and Digestive and Kidney Diseases, all patients here are females at least 21 years old of Pima Indian heritage. Get the Diabetes dataset here.
#load data and assigns it to a dataFrame(df)
df = pd.read_csv('diabetes.csv')
# Features:
- Glucose Level
- Blood Pressure
- Insulin Levels
- BMI (Body Mass Index)
- Age
- Skin Thickness
- Number of Pregnancies
- Diabetes Pedigree Function (Genetic Risk Factor)
# Target Variable:
- Outcome (binary: 0 = No, 1 = Yes).
3. Data Overview
In this section, we take a general look at the overview of the dataset to understand its structure, size, and content. Key objectives include examining the data’s features, types, and summary statistics, as well as identifying any missing or duplicated values.
We achieve this through various exploratory commands, such as:
head()andtail()to view the first and last rows.sample()for random rows to check data consistency.columnsandshapeto understand feature names and dataset size.info()to explore data types and memory usage.duplicated()andisnull()to locate duplicates and missing values.describe()for descriptive statistics of numerical features.
This initial analysis helps set the stage for data cleaning and preprocessing
# head --> First 5(default) rows of dataframe(df)
df.head()
# tail --> Last 5(default) rows of df
df.tail()
# sample --> random 10 rows of df
df.sample(10)
# columns --> A look at the all the features column and outcome column
df.columns
# shape --> Total number (size) of df (rows, columns)
df.shape
# info --> summary info of the df
df.info()
# sum of duplicate values
df.duplicated().sum()
# sum of missing values in each column in a descending order
df.isnull().sum().sort_values(ascending = False)
# statistical summary of df
df.describe()
4. Data Cleaning
While the dataset initially appeared to have no missing values from the data overview, a deeper inspection revealed certain features with zero values that are clinically unrealistic, given their medical context. For example, features like glucose, blood pressure, or BMI should rarely, if ever, be zero. These anomalies indicate missing or incorrect data, which could negatively impact the model’s accuracy and reliability.

The data cleaning objective looked to:
Identify columns where zero values are inappropriate based on medical knowledge.
Replace these zero values with
NaNto properly represent missing data.Addressing missing values using appropriate imputation techniques, such as mean, median, or mode, depending on the feature.
# replace zero columns with appropriate nan value
zero_columns = ['Glucose', 'BloodPressure','SkinThickness', 'Insulin', 'BMI' ]
# Replace zeros with NaN
df[zero_columns] = df[zero_columns].replace(0, np.nan)
# Check for actual missing values
df.isnull().sum().sort_values(ascending=False)


This function ensures that missing values are imputed in a way that preserves the relationship between the features and the target variable (Outcome). By imputing missing values based on the median within each outcome group, the method accounts for potential differences in distributions between diabetic (Outcome = 1) and non-diabetic (Outcome = 0) individuals. This approach enhances data integrity while minimizing the risk of introducing bias.
def impute_missing_values(df):
"""
Impute missing values in a DataFrame based on the median of each column,
grouped by the 'Outcome' column.
Parameters:
df (DataFrame): The input DataFrame containing the data.
Returns:
DataFrame: The DataFrame with missing values imputed.
"""
# Loop through each column in the DataFrame, except 'Outcome'
for col in df.columns:
if col != 'Outcome':
# Group by 'Outcome' and compute the median for the current column
median_values = df.groupby('Outcome')[col].median()
# Impute missing values for Outcome = 0
df.loc[(df['Outcome'] == 0) & (df[col].isnull()), col] = median_values[0]
# Impute missing values for Outcome = 1
df.loc[(df['Outcome'] == 1) & (df[col].isnull()), col] = median_values[1]
return df
# Apply the function
df = impute_missing_values(df)
5. EDA and Feature Engineering
EDA helps uncover patterns, relationships, and trends in the data, providing a deeper understanding of the features and their interactions which can aid in creating new features. It also reveals potential issues like skewness, outliers, or correlations that may impact model performance.
5.1 Analyze individual feature distributions to understand their spread and skewness.

5.2 Use pairplots and scatterplots to explore relationships between features.

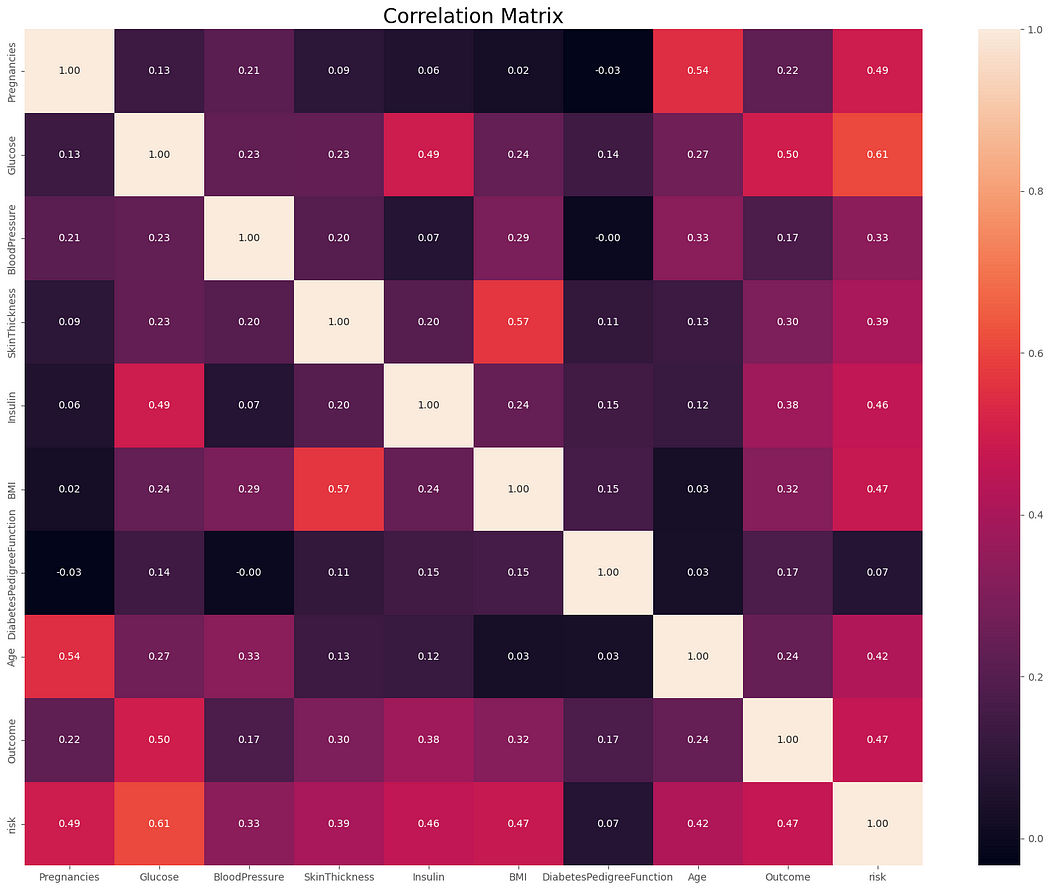
5.3 Examine the correlation matrix to identify significant associations between variables.
5.4 And Identify Outliers





Summary of Insights from Exploratory Data Analysis (EDA)
- Feature Distributions:
Most features show varying degrees of skewness. For example, features like BMI and Glucose exhibit distributions closer to normal, while others like Insulin show significant skewness.
Some features have outliers that may need further investigation or treatment to ensure model robustness.
2. Relationships Between Features:
Scatterplots and pairplots revealed interesting relationships. For instance, higher Glucose levels and BMI seem associated with diabetic outcomes.
Some features show clustering patterns corresponding to the diabetic (
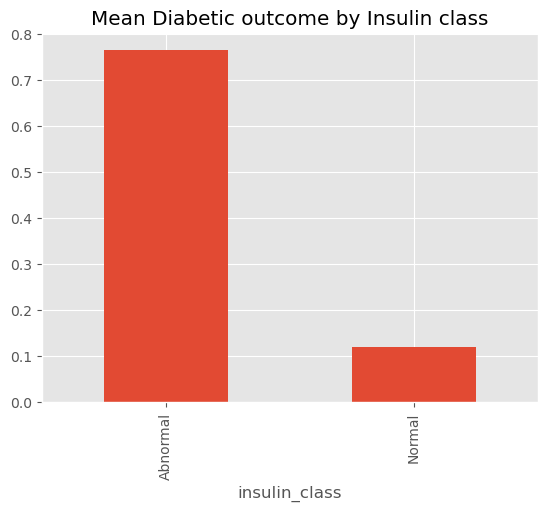
Outcome=1) and non-diabetic (Outcome=0) groups.Insulin Levels strongly correlated with diabetes. Individuals with abnormal levels above 166 showed a higher probability of diabetes.
Glucose Levels strongly correlated with diabetes. Individuals with glucose levels above 100 mg/dL showed a higher probability of diabetes.
Higher BMI values significantly increased diabetes risk, emphasizing the role of weight management.
Older individuals had a higher prevalence of diabetes.
With respect to gestational diabetes, those who have been pregnant more than 5 times had the highest probability of diabetes
3. Correlation Analysis:
Strong positive correlations were observed between features like Glucose and Outcome, indicating their importance for prediction.
Weak or negligible correlations for some features (e.g., Skin Thickness) suggest they might be less informative individually.
4. Outliers and Missing Values:
Certain features, like Insulin and Skin Thickness, have unrealistic zero values that were addressed in the data cleaning phase.
Outliers were clipped to either the upper or lower quartile
5. Class Imbalance:
- The dataset isn’t balanced, with the diabetic class being underrepresented.
6. New Features Created (Feature engineering)
Several new features were derived from the original dataset to enhance the model’s ability to capture patterns and relationships in the data:
BMI Class: Categorized BMI into meaningful bins based on standard medical classifications.
Insulin Class: Grouped insulin levels into classes to provide insight into insulin patterns.
Glucose Class: Categorized glucose levels to represent a patient’s risk category more effectively.
This analysis guides feature selection and informs preprocessing and modeling decisions.
6. Data Preprocessing
One-Hot Encoding: Categorical features such as bmi_class, insulin_class, and glucose_class were one-hot encoded to convert them into numerical representations suitable for machine learning algorithms. This ensures that these categories are treated independently and equitably during model training.
Data Scaling: To handle differences in feature scales, the dataset was divided into numerical and categorical subsets. Only the numerical data was scaled, as scaling one-hot-encoded categorical data can distort their representation.
The Robust Scaler was chosen for scaling the numerical data due to its robustness to outliers, ensuring the scaled values are not overly influenced by extreme values. After scaling, the numerical and categorical dataframes were concatenated back together, forming the final processed dataset. The data was then split into training and testing sets in an 80:20 ratio, preparing it for model development and evaluation.
This preprocessing approach ensures a clean and well-prepared dataset, minimizing the risk of biases and preserving data integrity for machine learning.
7. Model Development and Training
# List models
models = {
'Logistic Regression': LogisticRegression(random_state=42),
'Random Forest': RandomForestClassifier(random_state=42),
'Support Vector Machine': SVC(random_state=42),
'Decision Tree': DecisionTreeClassifier(random_state=42),
'K-Nearest Neighbour': KNeighborsClassifier(),
'Naive Bayes': GaussianNB(),
'Gradient boosting': GradientBoostingClassifier(random_state=42),
}
# Dictionary to store results
model_results = []
for name, model in models.items():
print(f'\n--- Evaluating {name} ---')
#Cross validation prediction
cv_scores = cross_val_score(model, X_train, Y_train, cv=10, scoring='accuracy', n_jobs=-1)
y_pred_cv = cross_val_predict(model, X_train, Y_train, cv=10, n_jobs=-1)
# Fit model on Train data
model.fit(X_train, Y_train)
y_pred_test = model.predict(X_test)
# CV Metrics
cv_mean_accuracy = cv_scores.mean()
cv_conf_matrix = confusion_matrix(Y_train, y_pred_cv)
cv_precision = precision_score(Y_train, y_pred_cv, average='weighted')
cv_recall = recall_score(Y_train, y_pred_cv, average='weighted')
cv_f1 = f1_score(Y_train, y_pred_cv, average='weighted')
cv_classification = classification_report(Y_train, y_pred_cv)
# Test Set Metrics
test_accuracy = accuracy_score(Y_test, y_pred_test)
test_conf_matrix = confusion_matrix(Y_test, y_pred_test)
test_precision = precision_score(Y_test, y_pred_test, average='weighted')
test_recall = recall_score(Y_test, y_pred_test, average='weighted')
test_f1 = f1_score(Y_test, y_pred_test, average='weighted')
test_classification = classification_report(Y_test, y_pred_test)
# Save result
model_results.append({
'Model': name,
'Cross-Validation Accuracy': cv_mean_accuracy,
'CV Conf Matrix': cv_conf_matrix,
'CV Precision': cv_precision,
'CV F1 score': cv_f1,
'CV Recall': cv_recall,
'CV Report': cv_classification,
"Test Accuracy": test_accuracy,
'Test Conf Matrix': test_conf_matrix,
'Test Precision': test_precision,
'Test F1 score': test_f1,
'Test Recall': test_recall,
'Test Report': test_classification
})
# Print metrics
print(f'CV F1 Score :{cv_f1: .2f}')
print(f'CV Recall Score :{cv_recall: .2f}')
print(f'Test F1 Score :{test_f1: .2f}')
print(f'Test Recall Score :{test_recall: .2f}')
print('\n Test set classification report:')
print(test_classification)
I implored a variety of machine learning algorithms to predict diabetes and identified the best-performing model based on key evaluation metrics.
Algorithms Used:
Logistic Regression
Random Forest
Support Vector Machine (SVM)
Decision Tree
K-Nearest Neighbour (KNN)
Naive Bayes
Gradient Boosting
To ensure fairness and consistency, all models were trained and evaluated using the same training and test splits. The cross-validation technique was applied to provide a robust estimate of model performance, reducing the risk of overfitting to a single data split.
Key metrics, including recall, F1-Score, and accuracy, were used to evaluate model performance. Special emphasis was placed on recall, given the importance of minimizing false negatives in a clinical setting.
Through this comprehensive approach, the best-performing model was selected, balancing predictive power with clinical relevance.
8.Model Evaluation
Evaluating the performance of each model is critical to ensure that the chosen model not only predicts accurately but also aligns with the clinical needs of the problem. The following key metrics were used for this evaluation:
F1-Score: A harmonic mean of precision and recall, ensuring a balance between the two metrics, especially when the dataset is imbalanced.
Recall: Measures the ability to correctly identify diabetic patients (true positives). Given the health implications, recall was prioritized to minimize false negatives, ensuring that no high-risk patients are overlooked.
Accuracy: Reflects the overall correctness of the predictions but can be misleading in imbalanced datasets.
Precision: Assesses the proportion of correct positive predictions, ensuring relevance in identifying true diabetic cases.
Result
The Gradient Boosting Classifier outperformed other models across these metrics, achieving the highest recall and F1-Score, making it the best choice for this dataset. This ensures both clinical reliability and strong predictive capability.

Feature Importance
Understanding feature importance helps interpret the model’s decisions and ensures its clinical relevance. The Gradient Boosting Classifier provides insights into which features contribute the most to diabetes predictions:
Insulin: With a score of 0.683831, this feature had the highest influence on model predictions, aligning with its known clinical significance in diabetes.
Glucose: The second most impactful feature (0.119013), reinforcing its critical role in identifying abnormal blood sugar levels.
Age: Ranked third (0.056160), indicating the increased risk of diabetes with advancing age.
BMI: With a score of 0.041287, BMI highlights the link between obesity and diabetes risk.
These findings not only validate the model’s alignment with medical knowledge but also provide actionable insights for early intervention strategies focusing on these key predictors.
Challenges and Limitations
Data Imbalance: The dataset had more non-diabetic cases compared to diabetic ones.
Limited Features: Critical predictors such as dietary habits, physical activity, and genetic predisposition were missing, which could have provided a more comprehensive risk analysis for diabetes prediction.
Generalizability: The dataset comprises female patients from the Pima Indian heritage, limiting the model’s applicability to other populations. Validation on larger and more diverse datasets is necessary to ensure the model performs well across different demographics.
Future Directions
Expanding Features: Future iterations of this project could include lifestyle-related factors (e.g., dietary patterns, physical activity levels), genetic markers, and socio-economic data to enhance prediction accuracy and provide a more holistic analysis of diabetes risk.
Deployment: Building a user-friendly application or API for clinicians and individuals can bridge the gap between machine learning and real-world usability. This would allow healthcare professionals to leverage predictions effectively for early diagnosis and personalized interventions.
Conclusion
In this article, we explored the essential steps involved in a machine learning project. These steps include data collection, analysis, preprocessing, model development, evaluation, and ultimately, using the trained model to predict new data points. Throughout the process, various models were tested to identify the one that delivers the best performance. In this project, we saw the immense potential of machine learning in advancing healthcare, particularly in addressing the global challenge of diabetes. we leveraged the robust capabilities of the scikit-learn library, which provides a comprehensive suite of tools for implementing and fine-tuning machine learning models efficiently.
Beyond its predictive capabilities, the project demonstrates the critical importance of clean data, appropriate scaling, and rigorous evaluation in building reliable machine learning models. While the dataset’s limitations, such as imbalance and lack of diverse features, present areas for improvement, they also open avenues for further exploration.
I hope you learnt a new thing just like I learnt carrying out this project. If you have any questions or contributions, please let me know in the comment, thank you.
The Codes for this project can be found on GitHub: https://github.com/cemmanuelonyema/ML_Diabetes
